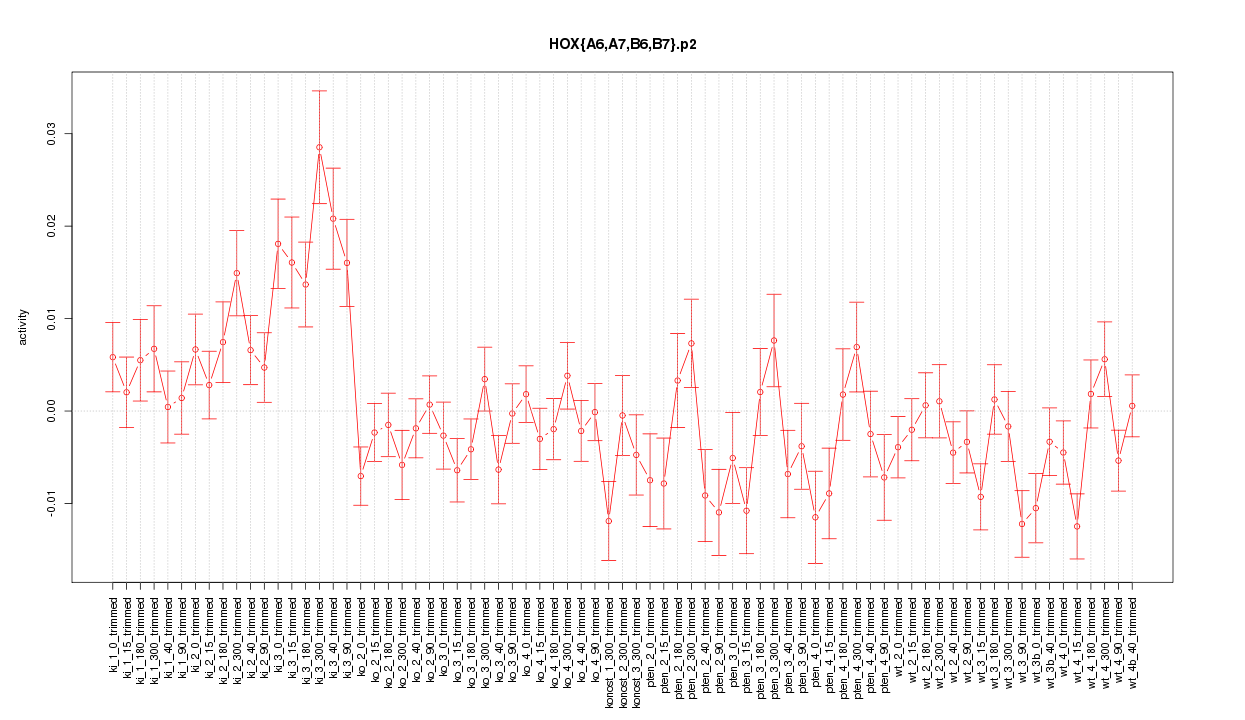
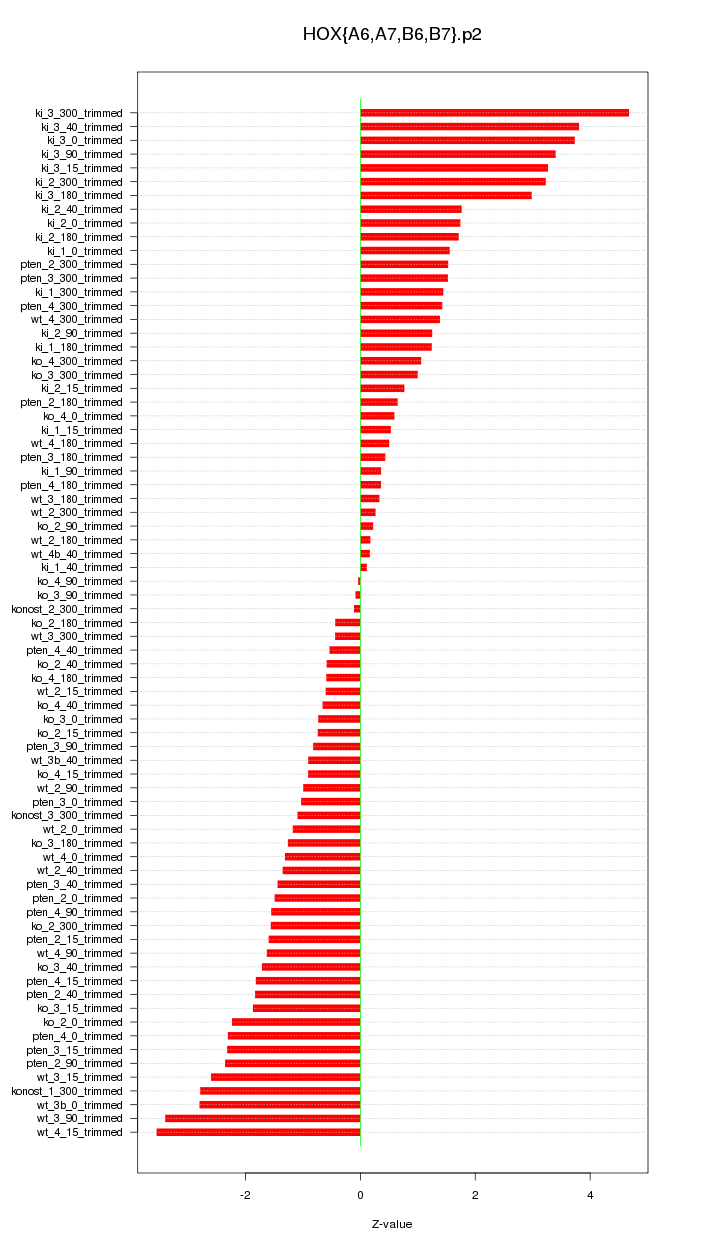
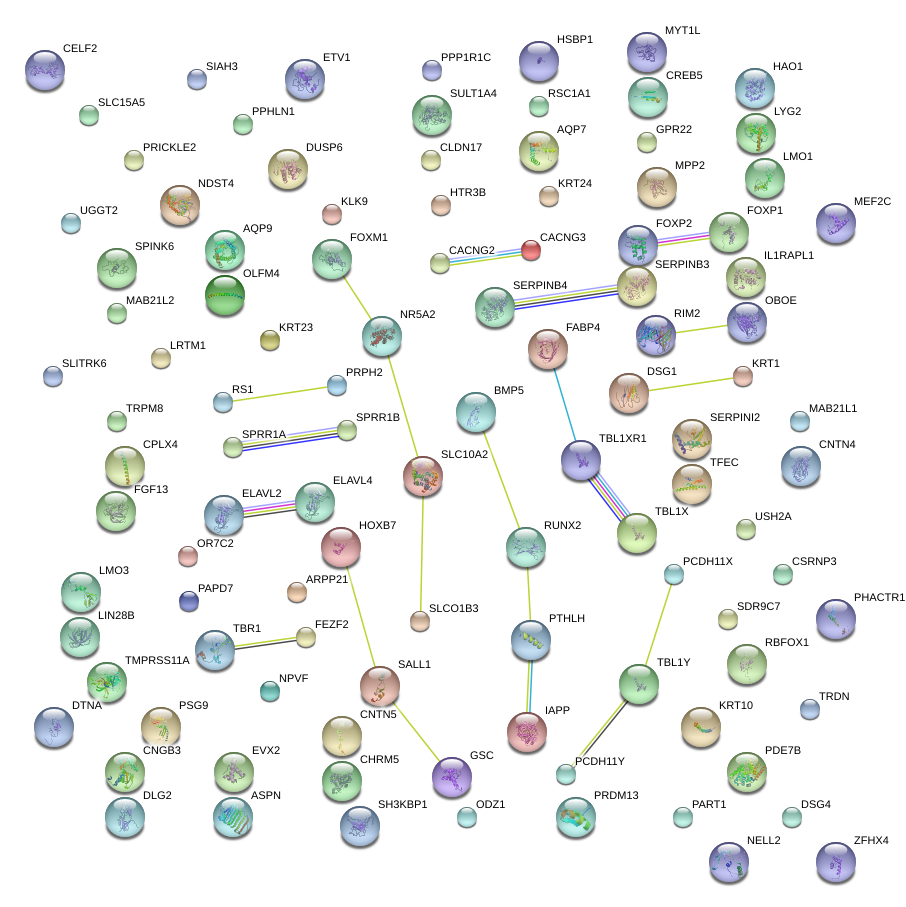
|
chr7_+_114066554
|
20.907
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr12_-_28124915
|
20.680
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr8_-_82395401
|
19.052
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_+_28452143
|
18.918
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_107110501
|
18.540
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr5_+_147582338
|
17.537
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr5_-_88119604
|
16.813
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr18_+_28898051
|
16.761
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr7_-_25268057
|
15.704
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr8_+_104831415
|
13.721
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr7_+_114055048
|
13.645
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr2_-_99871569
|
13.146
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr2_+_166428845
|
13.092
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr17_-_39093671
|
12.935
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_+_99426872
|
12.876
|
NM_175566
|
CNTN5
|
contactin 5
|
|
chr17_-_39092714
|
12.665
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr7_+_114055234
|
12.366
|
|
FOXP2
|
forkhead box P2
|
|
chr7_-_14025826
|
11.778
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr7_-_115670811
|
11.491
|
|
TFEC
|
transcription factor EC
|
|
chr11_+_113775517
|
11.435
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr7_+_114055171
|
11.351
|
|
FOXP2
|
forkhead box P2
|
|
chr2_+_162272619
|
11.176
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr12_-_16760935
|
10.907
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_176948689
|
10.880
|
NM_001080458
|
EVX2
|
even-skipped homeobox 2
|
|
chr6_+_105404922
|
10.831
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr5_+_59784004
|
10.684
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr7_-_115670774
|
10.630
|
|
TFEC
|
transcription factor EC
|
|
chr2_+_182850550
|
10.607
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr3_+_2553272
|
10.214
|
|
CNTN4
|
contactin 4
|
|
chr1_+_199996729
|
10.055
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr10_-_21807120
|
9.976
|
|
|
|
|
chr4_-_116034932
|
9.912
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr13_-_103719195
|
9.826
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr12_-_89745997
|
9.817
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr6_+_12716887
|
9.660
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr6_+_100054649
|
9.624
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr13_-_46425756
|
9.590
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr2_+_162272892
|
9.580
|
|
TBR1
|
T-box, brain, 1
|
|
chr12_-_16430618
|
9.567
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr12_-_89746244
|
9.354
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_-_38859976
|
9.353
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr13_-_86373328
|
9.336
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr17_-_38978824
|
9.332
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr12_+_20963637
|
9.221
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr15_+_58430394
|
9.189
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr6_+_12717036
|
9.120
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr2_+_226265601
|
9.119
|
NM_020864
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr1_+_152956563
|
9.066
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr2_+_162272800
|
8.989
|
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_115670828
|
8.853
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr14_-_95236351
|
8.851
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr19_-_51512803
|
8.831
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr18_-_56985693
|
8.831
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr2_+_234826042
|
8.779
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr12_-_57328044
|
8.743
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr17_-_46687925
|
8.733
|
|
HOXB7
|
homeobox B7
|
|
chr7_+_28475233
|
8.669
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chrX_-_19817907
|
8.652
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr22_-_37098620
|
8.633
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chrX_-_137821514
|
8.617
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr13_-_36050758
|
8.613
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chrX_-_124097601
|
8.569
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr12_+_89744714
|
8.561
|
|
LOC100131490
|
uncharacterized LOC100131490
|
|
chr4_-_41884568
|
8.545
|
|
|
|
|
chrX_+_28605558
|
8.541
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr3_-_64211111
|
8.516
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr12_-_45307710
|
8.481
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr21_-_31538970
|
8.476
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr1_+_50569581
|
8.427
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_-_71353910
|
8.408
|
NM_001244812
|
FOXP1
|
forkhead box P1
|
|
chr2_-_2334887
|
8.385
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_51185150
|
8.354
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr7_+_28725597
|
8.301
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_+_7382750
|
8.276
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr18_-_61329113
|
8.176
|
NM_006919
|
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
|
|
chrX_+_9431314
|
8.161
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_-_7921068
|
8.106
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chrX_+_91090459
|
8.089
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr12_+_21525801
|
8.040
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr12_-_89746238
|
7.981
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr5_-_88199868
|
7.965
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr11_-_84634219
|
7.955
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr17_-_46688279
|
7.923
|
|
HOXB7
|
homeobox B7
|
|
chr8_-_87755880
|
7.882
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chrX_-_19817854
|
7.880
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr13_+_53602829
|
7.876
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr6_+_45296053
|
7.870
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr3_-_167191808
|
7.756
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr3_+_35721115
|
7.747
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr6_-_55740387
|
7.712
|
|
BMP5
|
bone morphogenetic protein 5
|
|
chr10_+_11047339
|
7.695
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_-_71294229
|
7.693
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr6_+_136172802
|
7.691
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr1_-_216596737
|
7.511
|
NM_007123
NM_206933
|
USH2A
|
Usher syndrome 2A (autosomal recessive, mild)
|
|
chr12_-_53074172
|
7.471
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr8_+_77593506
|
7.426
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr9_-_95244750
|
7.250
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr15_+_34261088
|
7.217
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr6_-_123957941
|
7.210
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr3_-_62359189
|
7.204
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr6_-_55740358
|
7.177
|
NM_021073
|
BMP5
|
bone morphogenetic protein 5
|
|
chr18_+_28956739
|
7.171
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr18_+_32173281
|
7.166
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_-_95244634
|
7.114
|
|
ASPN
|
asporin
|
|
chr3_-_54961961
|
7.046
|
NM_020678
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1
|
|
chr4_-_68829143
|
7.043
|
NM_001114387
NM_182606
|
TMPRSS11A
|
transmembrane protease, serine 11A
|
|
chrX_-_18690187
|
7.042
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr8_+_77593522
|
7.033
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr4_+_88720701
|
7.003
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr8_+_75736771
|
6.923
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr12_-_99548867
|
6.910
|
NM_001204067
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_-_95166827
|
6.838
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr3_+_2280512
|
6.837
|
NM_001206955
|
CNTN4
|
contactin 4
|
|
chr3_-_114478117
|
6.832
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_145252530
|
6.794
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr6_-_108145507
|
6.774
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chrX_-_64196268
|
6.751
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr1_+_50574601
|
6.743
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr18_-_3874252
|
6.726
|
NM_001242764
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr5_-_134914968
|
6.715
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr7_+_39017608
|
6.681
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr13_-_84456473
|
6.678
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr5_+_170736287
|
6.603
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr6_+_153019029
|
6.595
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr1_+_198608216
|
6.569
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr17_-_10452939
|
6.555
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr5_-_76934373
|
6.548
|
|
OTP
|
orthopedia homeobox
|
|
chr12_-_28122884
|
6.499
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr9_+_118916069
|
6.463
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr21_-_39870303
|
6.435
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_14132920
|
6.412
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr2_-_233641225
|
6.406
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr11_-_36619776
|
6.374
|
NM_000536
NM_001243785
NM_001243786
|
RAG2
|
recombination activating gene 2
|
|
chr5_-_88179278
|
6.374
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_72649734
|
6.372
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr4_+_15341559
|
6.342
|
NM_001135170
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr14_+_76837689
|
6.310
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr13_-_84456424
|
6.305
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr3_-_74570262
|
6.301
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr10_-_118897811
|
6.274
|
NM_001112704
NM_199131
|
VAX1
|
ventral anterior homeobox 1
|
|
chr14_-_101036130
|
6.257
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr12_-_89745753
|
6.253
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr1_+_177140062
|
6.236
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr5_+_60933635
|
6.179
|
NM_173667
|
C5orf64
|
chromosome 5 open reading frame 64
|
|
chr1_+_50574589
|
6.154
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr10_+_11047274
|
6.142
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_174451356
|
6.134
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr4_+_88529680
|
6.133
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr5_-_76934508
|
6.108
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr2_-_29297126
|
6.101
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr9_+_27109138
|
6.070
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr18_-_31628557
|
6.066
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr10_-_50599906
|
6.055
|
NM_001080520
|
DRGX
|
dorsal root ganglia homeobox
|
|
chr2_-_2335080
|
6.038
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr8_-_42234562
|
6.034
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr5_+_161494647
|
6.022
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr5_+_36606667
|
5.962
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_+_80838125
|
5.904
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr1_-_217250230
|
5.896
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr21_-_40033617
|
5.893
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr6_+_27925018
|
5.892
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr5_+_36606456
|
5.888
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr13_-_103054027
|
5.852
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr6_+_54173202
|
5.846
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr5_-_141993691
|
5.832
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr4_-_57547845
|
5.824
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr19_-_51522953
|
5.823
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr1_-_197036350
|
5.800
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr3_-_97754147
|
5.757
|
NM_001105580
|
GABRR3
|
gamma-aminobutyric acid (GABA) receptor, rho 3
|
|
chr7_-_37025696
|
5.731
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_-_114429996
|
5.718
|
|
BCL2L15
|
BCL2-like 15
|
|
chr1_+_50571963
|
5.690
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr18_-_3845295
|
5.671
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr9_-_124989755
|
5.665
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr18_-_64271215
|
5.652
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr18_+_32073245
|
5.630
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr16_+_2653350
|
5.627
|
|
LOC652276
|
potassium channel tetramerisation domain containing 5 pseudogene
|
|
chr5_-_131879172
|
5.608
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr20_+_62795826
|
5.602
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr10_+_11207053
|
5.584
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr17_-_46671322
|
5.560
|
|
HOXB5
|
homeobox B5
|
|
chr20_-_50418946
|
5.554
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr12_-_52946734
|
5.531
|
NM_033448
|
KRT71
|
keratin 71
|
|
chr17_-_39123137
|
5.525
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr4_-_44653657
|
5.524
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr11_+_15133969
|
5.510
|
NM_001031853
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr3_+_132318980
|
5.498
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chrX_+_115567746
|
5.447
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr17_-_10421858
|
5.441
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chrX_-_110513750
|
5.438
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr14_+_62462540
|
5.433
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr16_+_2732475
|
5.425
|
NM_018992
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr3_-_114343052
|
5.405
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_21792815
|
5.392
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr11_-_84634437
|
5.367
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_-_112727778
|
5.359
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr10_-_69455926
|
5.331
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr3_+_127641901
|
5.320
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr12_-_70093064
|
5.312
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr3_+_173116243
|
5.307
|
NM_014932
|
NLGN1
|
neuroligin 1
|